Quick-Neuron™ Motor – Human iPSC-Derived Neurons (Healthy Donor)
Our proprietary transcription factor-based stem cell differentiation method produces neurons without a genetic footprint. Quick-Neuron™ Motor – Human iPSC-derived Neurons display typical neurite outgrowth and express a variety of neuronal markers, such as the pan-neuronal marker TUBB3 (red) and the motor neuron marker HB9 (green). When thawed and maintained according to the instructions in the user guide, the iPSC-derived neurons are viable long-term and are suitable for a variety of characterization and neurotoxicity assays.
Original price was: $729.00.$350.00Current price is: $350.00.
Advantages of iPSC-Derived Motor Neurons
~ 1 Week Differentiation
Functionally Validated
Highly Pure Population
No Genetic Footprint
Patient-Derived Motor Neurons
We offer motor neurons derived from human iPSCs licensed through The California Institute for Regenerative Medicine (CIRM). These neurons have a diverse genetic background, as hiPSC lines from over 1,500 donors are available in the CIRM repository. If you require neurons from a specific donor, contact us to discuss your needs.
Motor Neuron Protocol
Explore our detailed differentiation protocols, a step-by-step guide designed to simplify and optimize your laboratory procedures using our iPSC-derived cells and differentiation kits. These protocols leverage the latest advancements in iPSC technology to ensure efficient and reproducible results.


Motor neuron morphology is confirmed via phase contrast imagery: Representative images of Quick-Neuron™ Motor – Human iPSC-derived Neurons on days 1-7 post-thaw (scale bar = 100 μm).
Motor Neuron Characterization
Motor Neuron Marker Expression
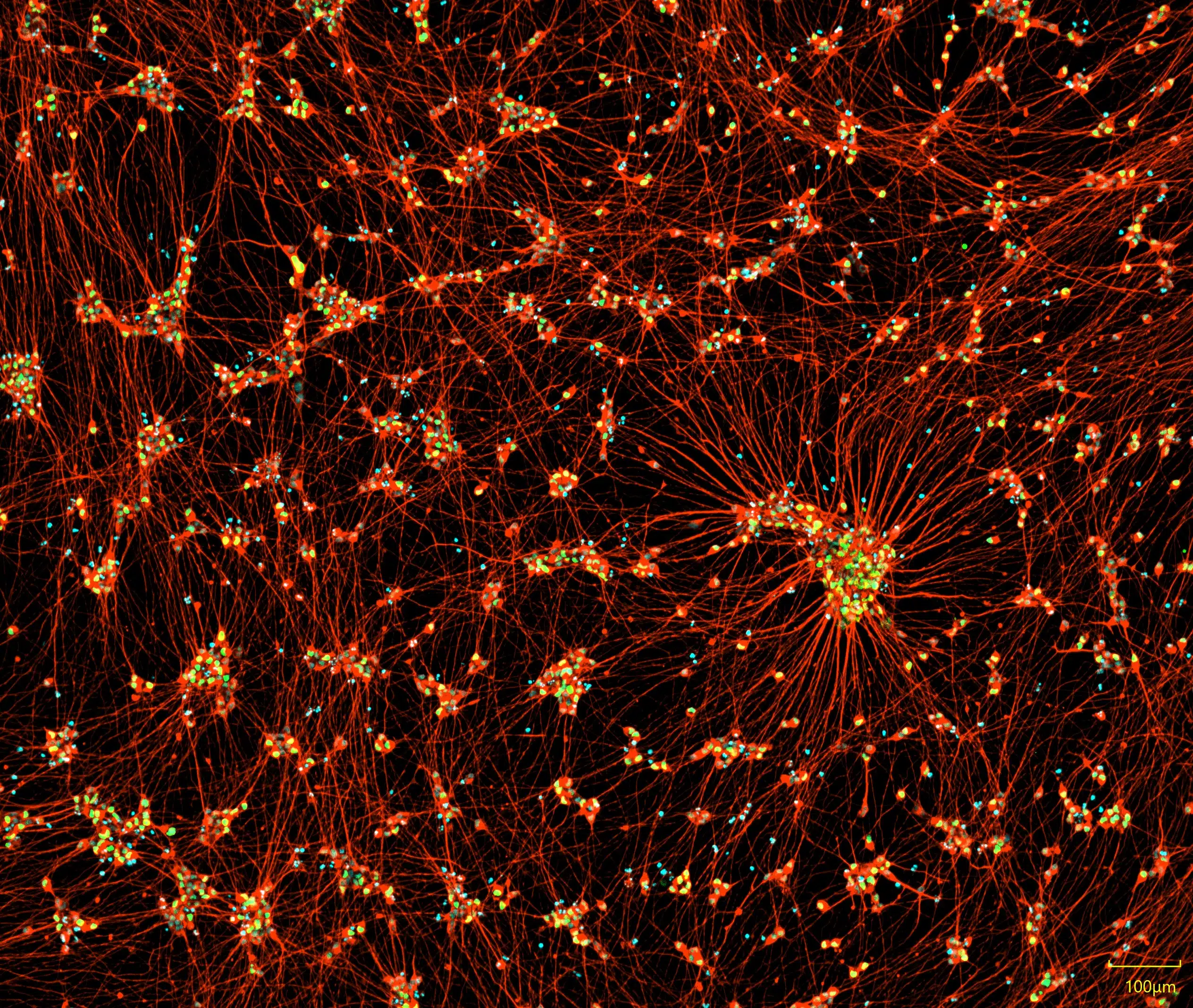
Merge

TUBB3

HB9

Nuclei
iPSC-derived motor neurons express neuronal markers and display typical neurite outgrowth. Immunofluorescent staining of iPSC-derived motor neurons at day 10 post-differentiation. Cells exhibit extensive neurite outgrowth and co-expression of the pan-neuronal marker TUBB3 and the motor neuron-specific marker HB9.
Real-time Quantitative PCR Analysis

Real-time quantitative PCR analysis of expression levels of genes CHAT, HB9, and ISL1 were examined. The graph shows gene expression in Quick-Neuron™ Motor – SeV culture on day 10 post-differentiation. The relative gene expression is normalized to phosphoglycerate kinase 1 (PGK1), and then calculated as a fold induction relative to undifferentiated hPSCs as a control. Error bars show standard deviation.
TDP-43 Mislocalization in ALS Motor Neuron Models
TDP-43 mislocalization is a defining molecular feature of amyotrophic lateral sclerosis (ALS). Our ALS motor neuron models enable detailed investigation of TDP-43 biology and support a range of applications in disease mechanism studies and therapeutic discovery.
Quantification of TDP-43 localization in Quick-Neuron™ motor neurons. Violin plots show the ratio of cytoplasmic to total TDP-43 puncta in cultures derived from a sporadic ALS patient and a healthy control. Significance was quantified by T-test. Stars denote statistical significance: **** = p <0.0001.

Product Specifications
| Parameters | Specifications |
|---|---|
| Product Name | Quick-Neuron™ Motor - Human iPSC-Derived Neurons |
| Catalog No. | MT-SeV-HC-CW50065 |
| Product Components | Cryopreserved cells, Component N1, Component A, Component P, and Component K |
| Starting Material | iPSCs derived from peripheral blood mononuclear cells (CIRM line CW50065) |
| Storage Conditions | Frozen cells should be stored in liquid nitrogen (vapor phase). The rest of the components should be stored at -20°C. |
| Cell Type | Motor Neurons |
| Culture Type | Feeder Cell-Free |
| Disease | Healthy Control |
| Donor Sex | Female |
| Donor Age At Sampling | 74 |
| Donor Race Ethnicity | Caucasian, not Latino |
| Patient History | See Resources section for more information. |
| Reprogramming Method | Episomal vector |
| Induction Method | Transcription factors delivered by Sendai virus |
| Growth Properties | Adherent |
| Number of viable cells | > 1.0 million viable cells per vial upon thawing |
| Cell viability and remaining live cells | >50% at day 1, >211 live cells per mm2 >50% at day 7, >211 live cells per mm2 |
| Differentiation | >80% TUBB3 positive cells >50% ChAT positive cells among TUBB3 positive cells >40% HB9 positive cells among TUBB3 positive cells |
| Sterility | No growth observed for Bacteria and Fungus |
| Mycoplasma | No contamination detected |
| Morphological Observation | Cells are adherent and neurites exhibit substantial outgrowth, elongation and branching, indicative of a differentiated phenotype. |
| Restricted Use | For Research Use Only. Not for use in diagnostic or therapeutic procedures. |
Resources
Quick-Neuron™ Motor – Human iPSC-Derived Neurons
Quick-Neuron™ Motor – SeV Kit
Human Motor Neurons With SOD1-G93A Mutation Generated From CRISPR/Cas9 Gene-Edited iPSCs Develop Pathological Features of Amyotrophic Lateral Sclerosis.
Modeling ALS Using Patient-Derived iPSCs: A Human-Relevant Platform for Disease Research and Therapeutic Discovery (SfN 2025)
Related Products
FAQs
What kind of transcription factors are used for differentiation induction?
It is a proprietary formulated RNA and cannot be disclosed.
Why do I see many floating cells and/or cellular debris on day 1?
hPSCs were likely damaged while they were harvested. hPSCs may have been mechanically harvested by scraping or were pipetted too much or too vigorously. Lift Solution D1-treated hPSCs only by pipetting them up and down up to 15 times. The mixing of hPSCs with SeV may have also been done too strongly. Gently rock the plate several times to mix hPSCs with SeV.
What should I do if I find on day 1 that cells were accumulated in the center of the well?
It is not recommended to start mRNA-based differentiation induction because the differentiation efficiency is expected to be quite low. For successful differentiation using our mRNA-based methods including Mesendoderm Booster, hPSCs should be evenly distributed in the well. It is likely that the plate was shaken too much, and cells were accumulated in the center of the well while they were floating. Next time, please handle the plate more gently or it can be left on a flat bench top without vibration for 15 min before it is put inside a CO₂ incubator.
What should I do if I find on day 1 that my culture was seeded only in the center but not in the periphery of the well?
Unless cells are accumulated in the center of the well, the instructions can be followed since this is an issue of coating. We have observed that 4-well plates available from Nunc sometimes do not allow cells to settle down in the periphery of the well when our coating conditions are applied. Next time, please use a 24-well plate and try the following: 1) incubate the plate with Coating Material A at 4°C overnight instead of 37°C for 2 hours, 2) do not aspirate the supernatant completely or leave the supernatant just enough to cover the well bottom surface after coating with Coating Material A is completed and 3) do not touch the bottom of wells with fingers when handling the plate.
I plated cells as instructed, so why does my culture look less confluent than the one shown in the user guide on day 1?
hPSCs were likely damaged while they were harvested. Ideally, hPSCs should be lifted only by pipetting several times using Solution D1 as instructed in the user guide. Even if pipetting 15 times does not lift all of the cells in the culture, do not attempt to do more pipetting but collect only cells that come off. Instead rinse the culture with PBS and start Solution D1 treatment again but with longer incubation (up to 20 minutes). Next time please reduce the concentration of the substrate used to coat the dish/well for the maintenance of the source hPSCs to 50-70%.
Why does my culture have colonies that appear balled up and round on day 1?
It could be that Coating Material A was diluted at room temperature or diluted using PBS warmed to room temperature. Keep both Coating Material A and PBS on ice before dilution.
Can I use an orbital shaker for SeV infection while incubating cell suspension for 10 minutes?
The purpose of flicking the tube containing cells and SeV every 2 minutes during the 10 minute incubation is to prevent cells from settling down and forming aggregates. If the orbital shaker can do so, then it is possible to use it.
Can I use Geltrex or Matrigel as a coating substrate for hPSC cultures?
Our kits are not optimized with these substrates. However, if your hPSC differentiation protocols are already optimized with such a substrate, you may try using one of these with our kit at your own risk.
I was able to harvest more hPSCs than I needed for the kit. Can I replate or freeze the leftover hPSCs to maintain their culture?
No. If you’re following the instructions given in the user guide, the hPSCs will be harvested in a differentiation medium so the leftover hPSCs cannot be maintained.
Why are there many aggregates after harvesting hPSCs?
After harvesting hPSCs, cells should not be left in the collection tube for more than 5 minutes. Leaving cells for a longer period may cause the formation of cellular clumps which interferes with infection. Single cells are best for infection.
Why doesn’t Solution D1 dissociate my hPSC culture without scraping?
The concentration of the substrate used to coat the dish/well was likely too high, so Solution D1 treatment requires longer incubation than 10 minutes (up to 20 minutes). Next time, please reduce the concentration of the substrate to 50-70%.
Why do differentiation kits require hPSCs to be maintained under StemFit or StemFlex conditions?
hPSCs should be adapted to passaging as single cells prior to differentiation. Our protocols have only been tested with StemFit/StemFlex, so we can’t guarantee results using other culture conditions.
How long do I need to maintain hPSCs prior to beginning to use your kit?
Before beginning differentiation, cultures should be healthy and displaying typical morphologies (i.e., round colonies with tightly packed cells exhibiting a small cytoplasmic area), few spontaneously differentiating cells, and normal growth rates (i.e., doubling once a day). We recommend growing the cells for at least 14 days post-thaw before differentiating and not allowing the cultures to reach more than 80% confluency.
Do I need a license agreement for any of Ricoh Biosciences’ products?
No. You don’t need any licence or material transfer agreement (MTA) to use our differentiation kits or iPSC-derived cells. However, please be advised that these products are for research use only.
Is Sendai virus (SeV) dangerous?
SeV is not pathogenic to humans (i.e., humans are not the natural host of the virus) and the infection does not persist in immunocompetent animals. Furthermore, SeV used in our kits does not produce infectious viral particles upon transduction to host hPSCs and is a temperature-sensitive mutant, such that it is active at 33℃ but becomes inactive at 37℃. However, because SeV can be transmitted by aerosol and contact with respiratory secretions and is highly contagious, appropriate care must be taken to prevent potential mucosal exposure to the virus. Our SeV-based kits must be used under Biosafety Level 2 (BL-2) containment with a biological safety cabinet or a laminar flow hood and with appropriate personal protective equipment. In the event that the virus comes into contact with skin or eyes, decontaminate the affected area by flushing with plenty of water and follow the safety manual prepared by your laboratory and approved by your Institutional Biosafety Committee.
Will SeV remain active after differentiation?
No. The SeV used in our kits is a temperature-sensitive mutant that is active at 33℃ but becomes inactive at 37℃, which is the temperature instructed in the user guides post-differentiation.
Does Quick-Tissue™ technology leave a genetic footprint?
Sendai virus (SeV) is an RNA virus, so it does not integrate into the genomic DNA. In principle, a foreign gene introduced intracellularly in the form of RNA is quickly translated and expressed because, unlike DNA, RNA does not need to enter the nucleus for forced expression, thereby providing no chance of mutagenesis. This is discussed in the following review paper: Yamamoto, et al., (2009) “Current prospects for mRNA gene delivery.” Eur. J. Pharm Biopharm 71, 484-489.
Contact Us
Have a question about our products, services, or custom projects? Our team is here to help—reach out and we’ll get back to you as soon as possible.
Subscribe
By signing up you are agreeing to our Privacy Policy

